SightBot: ChatGPT-powered research insights with PubMed citations
Nikita Kedia & Suvansh Sanjeev • 2023-04-03
Check out our research chatbot demo and learn how Brilliantly can boost your business with AI
For a limited time, try SightBot without having to generate an API key!
We are thrilled to introduce SightBot, an AI-powered chatbot tailored to meet the requirements of the biomedical research community! With its natural language interface, you can converse with SightBot to navigate through literature and receive citations from peer-reviewed sources. Brilliantly empowers clients across industries to deploy custom AI solutions for their use case, and we’re excited to share this demo as an example of what we can build for you.
Find the code for SightBot at these repos: frontend, backend.
Feature Overview
Key features of SightBot that make it an exciting tool for biomedical researchers:
- Follow-ups: we built a module that allows SightBot to handle both follow-ups and new questions in subsequent messages, creating a seamless and natural user experience.
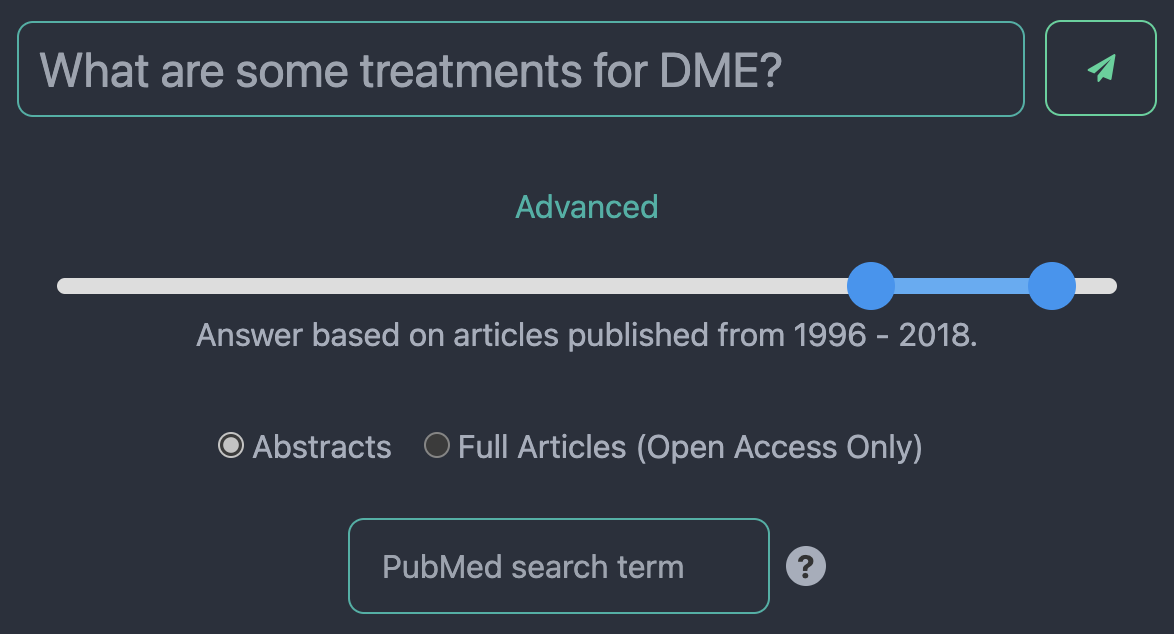
- Document type setting: you can choose whether questions are answered from abstracts of all PubMed articles or from the full article text of the open access subset of PubMed.
- Filter by year: you have the option to filter articles by year of publication, enabling more focused searches.
- PubMed search override: we automatically generate search terms to get relevant articles, but you can pick your own for finer-grained control.
- Export citations to BibTeX: we provide an option to copy citations to your clipboard for easier bibliography creation.
Fig. 1: The SightBot query options. Users can filter articles by publication year, choose whether to have the chatbot answer based on abstracts or full texts, and specify the search term sent to PubMed rather than relying on the SightBot-extracted search term from their overall query.
How SightBot Works
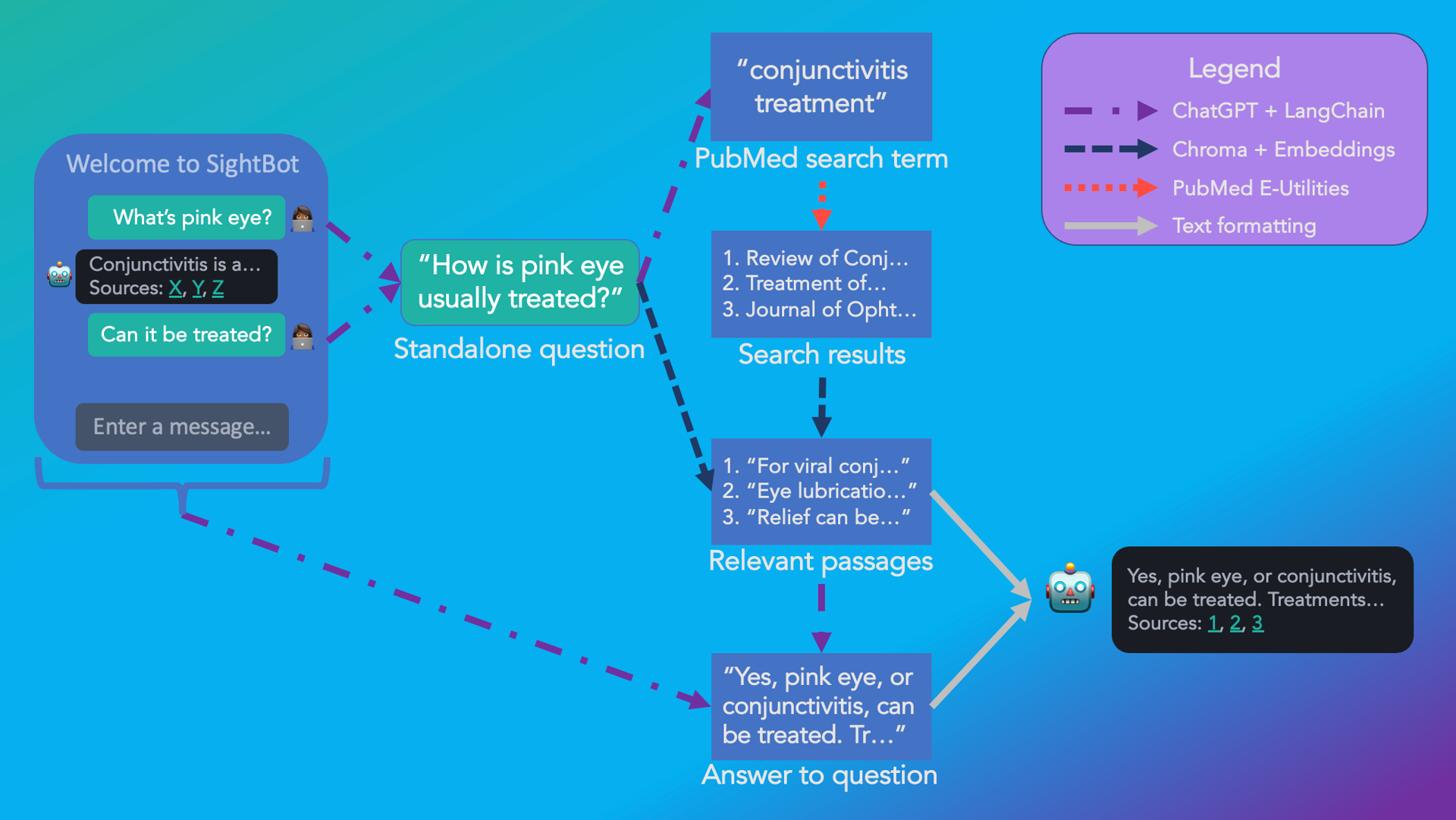
SightBot combines the powerful language understanding of ChatGPT with PubMed search to answer queries. Let’s break it down, step by step 😉
Fig. 2: The flow of information in SightBot from user message to a response, complete with citations.
1. Combine the user message with prior conversational context to produce a standalone question
The user’s natural expectation for a chatbot is to be able to retain conversational context for follow-up questions. For example, in the below conversation, the user’s follow-up is only answerable when paired with context from the original question.
Fig. 3: An example of a conversation with SightBot where follow-ups need to be combined with previous messages in order to be answerable. In this example, the combined question yielded by our module is “Does conjunctivitis affect the nose?” and the extracted search term is “conjunctivitis nose.”
Creating a PubMed search term from the second message alone would not yield useful documents to answer the question. Our search term extraction module pulls in relevant context from the conversational history to produce a fully-specified question.
2. Extract a search term from the question
In order to retrieve relevant documents to answer the question resulting from the previous step, we need to craft a search term for PubMed. Sticking to the theme, we once again use ChatGPT to get the job done, this time with a custom prompt (bolding added for readability):
Given a question, your task is to come up with a relevant search term that would retrieve relevant articles from a scientific article database. The search term should not be so specific as to be unlikely to retrieve any articles, but should also not be so general as to retrieve too many articles. The search term should be a single word or phrase, and should not contain any punctuation. Convert any initialisms to their full form. Question: What are some treatments for diabetic macular edema? Search Query: diabetic macular edema Question: What is the workup for a patient with a suspected pulmonary embolism? Search Query: pulmonary embolism treatment Question: What is the recommended treatment for a grade 2 PCL tear? Search Query: posterior cruciate ligament tear Question: What are the possible complications associated with type 1 diabetes and how does it impact the eyes? Search Query: type 1 diabetes eyes Question: When is an MRI recommended for a concussion? Search Query: concussion magnetic resonance imaging Question: <insert question here> Search Query:
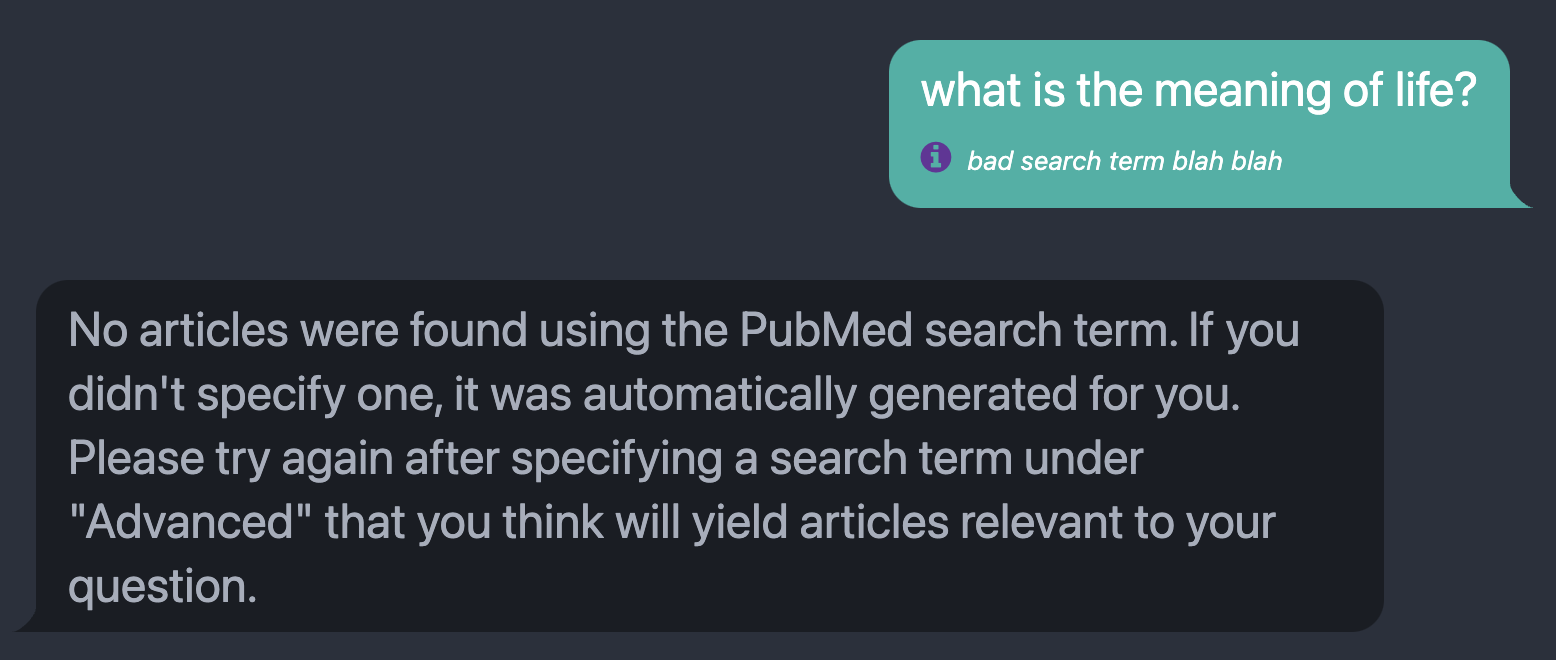
In our testing, ChatGPT performed fairly well in this task. However, there will inevitably be cases where the user’s query is niche or complex enough that this process fails, yielding a bad search term and no search results. To protect against this case, we display the automatically generated search term for each user message, as shown in Figure 4. The user can specify a PubMed search term that will override this automatic extraction process, as pictured in Figure 1.
Fig. 4: The message returned when the PubMed search returns no relevant articles, prompting the user to specify or modify the search term passed to PubMed to answer their query. For this example, the displayed search term next to the purple info icon was manually specified to elicit the error message.
3. Get relevant papers from PubMed
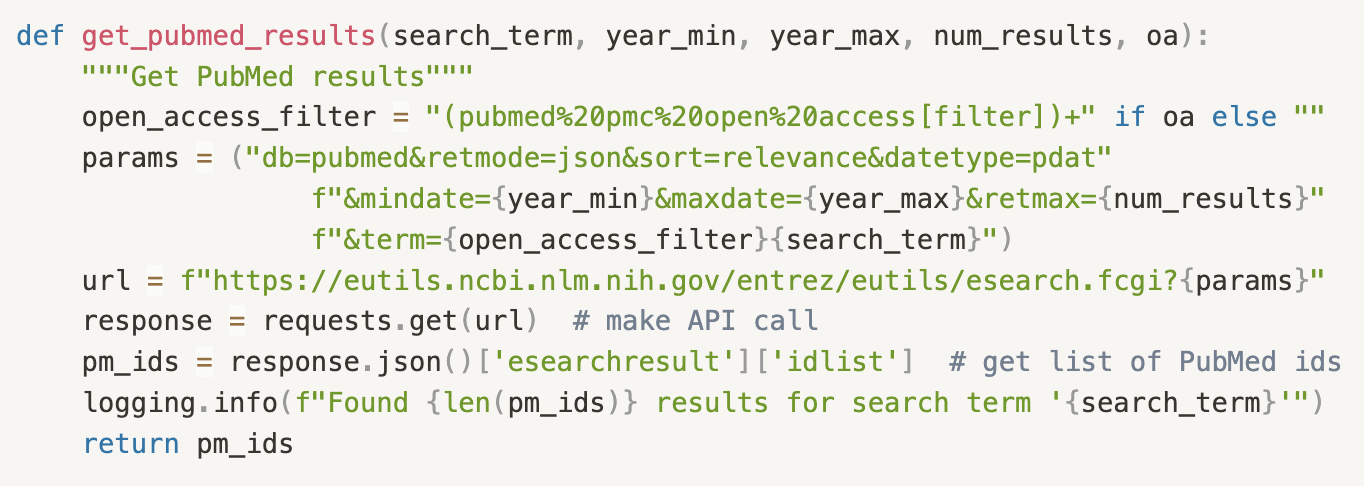
Having extracted the search term we need, we can now make use of the PubMed E-utilities and BioC API to automate a search over the expansive PubMed database of biomedical papers. If the user opts to search over abstracts (the default option), we are able to search over the full database. If the user chooses the full text option, we search over the open access subset (for which the full text is available through the API).
Fig. 5: A Python function to retrieve the most relevant article PubMed IDs for a given search term, filterable by publication year and optionally constrained to the open access subset, depending on whether we need to retrieve the associated full text or just the abstract. Not pictured is code to retrieve and parse the article/abstract text using the PubMed IDs.
4. Chunk and embed these documents into vectors
In order to figure out what parts of these documents we should pass off to ChatGPT to answer the final question, we need to be able to evaluate their relevance to the question. To do this, we embed them, or map them into vectors of numbers that represent the meaning of the text. We use OpenAI’s Embeddings API to accomplish this. Importantly, we break the documents up into chunks of smaller sizes. This way, we can eventually fit several of the most relevant chunks into the prompt for the final question. We use Chroma as our embedding vector database.
5. Embed the original query and retrieve the most similar passages
To retrieve the most relevant document chunks, we embed the user’s original question and pass this embedding vector to the Chroma database from the previous step. Chroma lets us retrieve the most similar text passages, as measured by cosine similarity of the corresponding embedding vectors.
6. Ask ChatGPT to answer the question based on these passages
Finally, we place the most relevant passages retrieved in the previous step into a single prompt along with the question from step 1 and obtain an answer from ChatGPT. The answer, the most relevant texts used to answer the query, and direct links to the articles are returned to the user, as pictured in Figure 3. We also provide an option to copy the associated BibTeX citations to the clipboard.
While SightBot may still suffer from occasional hallucinations inherent to the underlying ChatGPT model, the key advantage is that we provide citations. This allows you to verify information and delve deeper into the sources, ensuring a more accurate understanding.
Getting Started with SightBot
For a limited time, try SightBot without having to generate an API key!
To access SightBot, generate an OpenAI API key and begin using the chatbot to experience its utility for medical research. Each interaction with SightBot costs on the order of a penny (charged by OpenAI directly), making it an affordable solution for your research inquiries.
For ease of use, bookmark the URL
https://sightbot.brilliantly.ai?key=insert-openai-key-hereBrilliantly’s Custom AI Solutions
At Brilliantly, we offer a diverse range of services tailored to meet your evolving needs in the rapidly-changing world of AI.
- Custom chatbot solutions designed to work seamlessly with your specific documents
- Comprehensive AI consulting services designed to empower your business, providing valuable insights and strategies to navigate the AI landscape
- Executive coaching for forward-thinking leaders who aim to stay informed and ahead of AI trends, ensuring they are well-equipped to make informed decisions in an AI-driven world.
Let Brilliantly be your partner in navigating the exciting and dynamic realm of artificial intelligence. If you're interested in exploring AI solutions for your needs, we encourage you to get in touch with the Brilliantly team, either by filling out the relevant contact form on our website or by emailing us at [email protected].
Partnership and Future Research
SightBot was developed by Brilliantly’s Suvansh Sanjeev with Nikita Kedia, a medical student at the University of Pittsburgh. A review article and user study looking at the uses and impact potential for LLMs in ophthalmology is forthcoming, led by Kedia and Dr. Jay Chhablani of the University of Pittsburgh Medical Center, in partnership with Brilliantly.
There is plenty of room for optimization and improvement of the approach used here, some of which we will investigate in the future. Domain-specific custom embeddings, as used by the related BioMedLM, would likely represent a boost in embedding quality over the general OpenAI Embeddings we used, yielding higher quality semantic search over PubMed articles (Steps 4 and 5).
Hypothetical Document Embeddings, supported in LangChain, may also improve the semantic search performance over embedding the query directly. This approach involves generating fake but semantically plausible answers to queries, and retrieving relevant documents using the embedding of these fake answers, rather than of the queries themselves. The idea is that the “hypothetical documents” may share more similarities in embedding space with the truly relevant documents than the query alone.
In the process of generating this writeup, we encountered the wonderful Elicit, a free research assistant using LLMs to automate researcher workflows. Elicit streamlines many research processes ranging from literature reviews to idea generation, including many of the use cases we envisioned in building the SightBot demo. Highly recommend checking them out!
References / Software Used
See More Posts
brilliantly Blog
Stay connected and hear about what we're up to!